A Phylogeny of the Ixodes Genus (Hard-Bodied Ticks)
Jewel Voyer, Dr. Rachel Schwartz
The University of Rhode Island: Department of Biological Sciences
Abstract
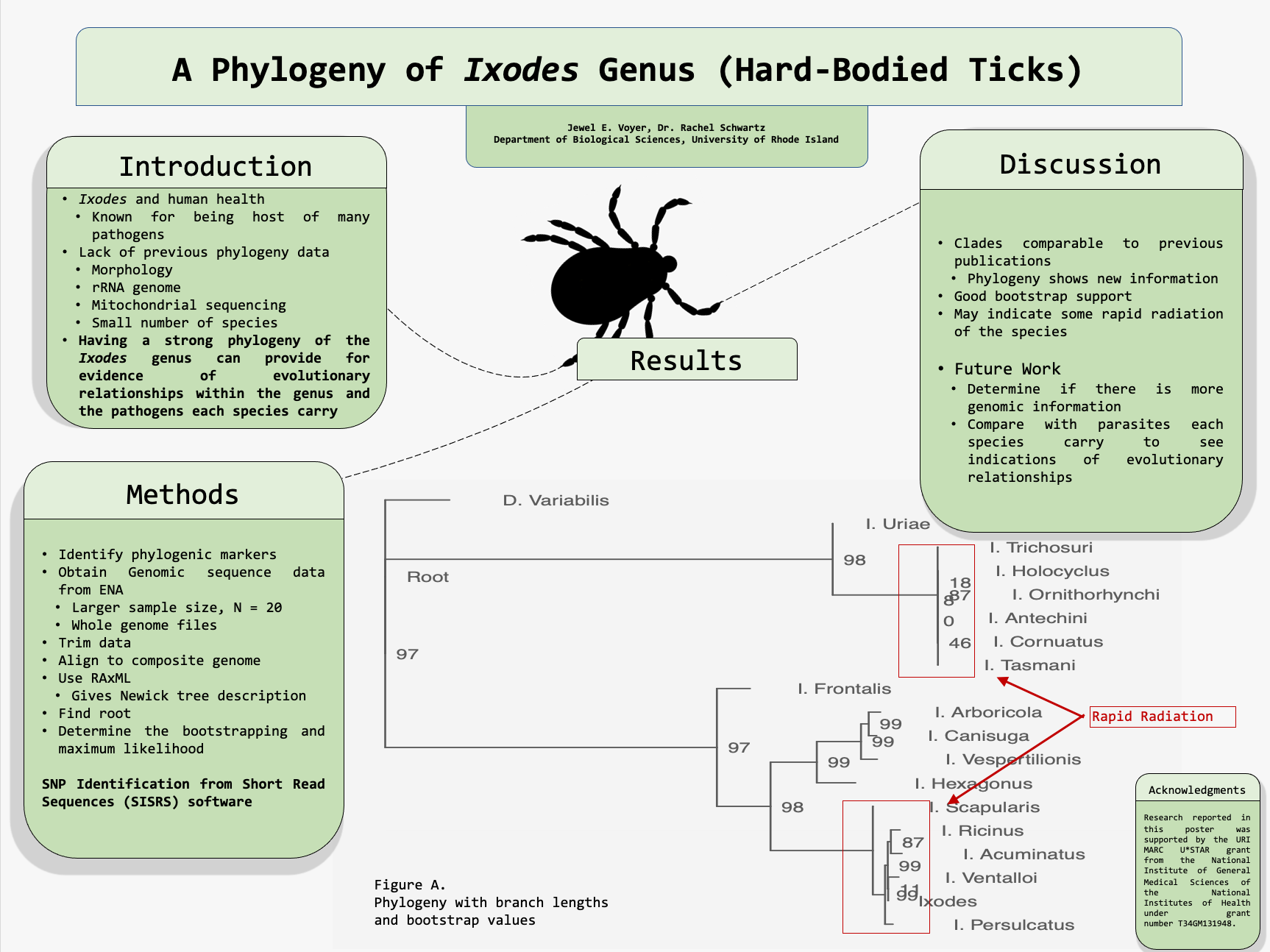
A phylogeny provides evolutionary relationships between certain taxa which then implements descendants of each taxon as well as contains the nodes representing the common ancestor. This phylogeny was created with the purpose of determining possible correlations between the species and human bacterial pathogens that they host. In order to do so, we will determine if the species closely related had similar bacterial pathogens compared to those which were farther away from each other on the phylogeny. This will be done by using the phylogeny and making a decision on whether each bacterial pathogen integrated with separate hard shell tick species who were more closely related versus if the bacterial pathogens were more generalized throughout each species in an incoherent order. The phylogeny provided a visualization of the relationships within the Ixodes genus which showed the evolutionary relationship between the species. We focused on what the phylogeny of the Ixodes genus and the human bacterial pathogens that each species hosts can inform us about the possible connections between this phylogeny and the pathogens. These connections will be determined from inspecting the phylogeny and mapping the relation between the hard-bodied tick species and then comparing the adjacent Ixodes tick’s relative pathogens to those in the remote location on the phylogeny to those ticks more distant on the phylogeny. We obtained raw, publicly available, genomic sequence data from the European Nucleotide Archive. We used these data in the SISRS software to identify phylogenetic markers. We then built phylogenies and examined support using maximum likelihood, bootstrapping, and site concordance factors in the software IQ-TREE. From there we will be able to conclude whether the bacterial pathogens were hosted on the hard-bodied ticks which were more closely related to the phylogeny or if the bacterial pathogens were hosted by ticks more broadly. We will gather data of the human bacterial pathogens each species carries from known, published, primary studies. The data will be used to generate evolutionary relationships between the Ixodes species and their known bacterial pathogens. This will allow us to see the progression of whether species relatively close on the phylogeny carry similar groups of bacteria to each other versus those that are farther away on the phylogeny.
Poster
Personal Video
Discussion
Please leave your comments on the following website:
https://showcasecomments.weebly.com/student-showcase-comments/discussion
Thank you!
